PySpark — The Famous Salting Technique
Out-of-memory errors are the most frequent and common error known to every data engineer. Data Skewness and Improper Shuffle are the most influencing reason for the same.
Before Spark 3 introduced — Adaptive Query Language(AQL), there was a famous technique called “Salting” which was used to avoid data skewness and distribute data evenly across partitions.
From Spark 3.2.0 AQL is enabled by default. Checkout documentation — https://spark.apache.org/docs/latest/sql-performance-tuning.html#adaptive-query-execution
Basically the technique involves adding a simple salt to the keys which are major part of joining datasets. Now, in place of the normal key, we use the salted key for join.
Currently this might sound a bit weird but the below example in action will clear all your doubts.
As always, we will start from - creating our Fact & Dimension datasets. Following is a Python code to generate random data for Fact & Dim
# Lets create the example dataset of fact and dimension we would use for demonstration
# Python program to generate random Fact table data
# [1, ,"ORD1001", "D102", 56]
import random
def generate_fact_data(counter=100):
fact_records = []
dim_keys = ["D100", "D101", "D102", "D103", "D104"]
order_ids = ["ORD" + str(i) for i in range(1001, 1010)]
qty_range = [i for i in range(10, 120)]
for i in range(counter):
_record = [i, random.choice(order_ids), random.choice(dim_keys), random.choice(qty_range)]
fact_records.append(_record)
return fact_records
# We will generate 200 records with random data in Fact to create skewness
fact_records = generate_fact_data(200)
dim_records = [
["D100", "Product A"],
["D101", "Product B"],
["D102", "Product C"],
["D103", "Product D"],
["D104", "Product E"]
]
_fact_cols = ["id", "order_id", "prod_id", "qty"]
_dim_cols = ["prod_id", "prod_name"]
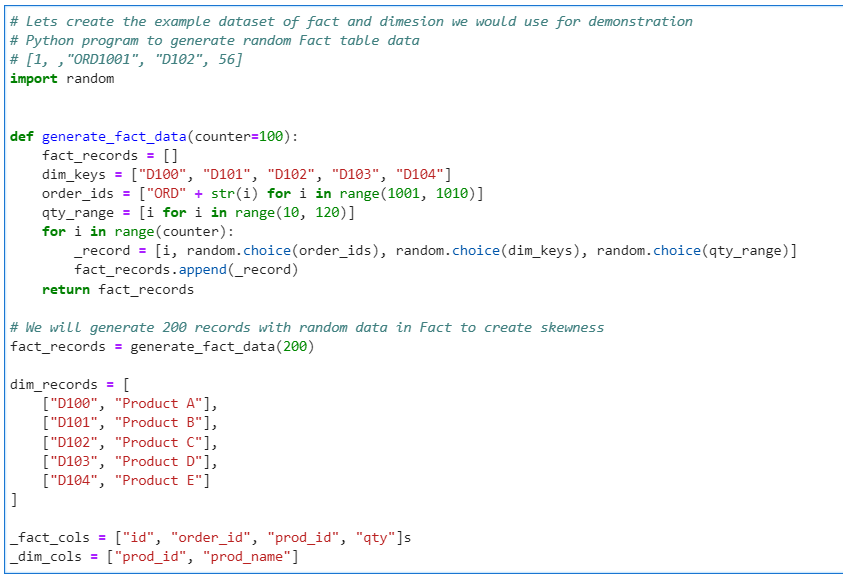
Generate the Fact Data Frame
# Generate Fact Data Frame
fact_df = spark.createDataFrame(data = fact_records, schema=_fact_cols)
fact_df.printSchema()
fact_df.show(10, truncate = False)
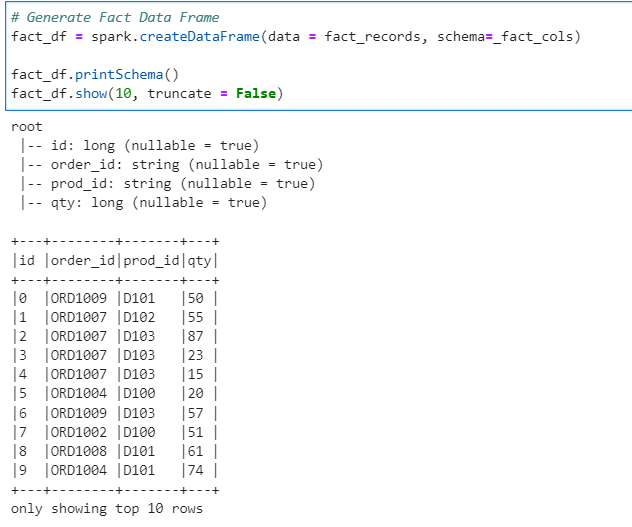
Generate Dimension Data Frame
# Generate Prod Dim Data Frame
dim_df = spark.createDataFrame(data = dim_records, schema=_dim_cols)
dim_df.printSchema()
dim_df.show(10, False)
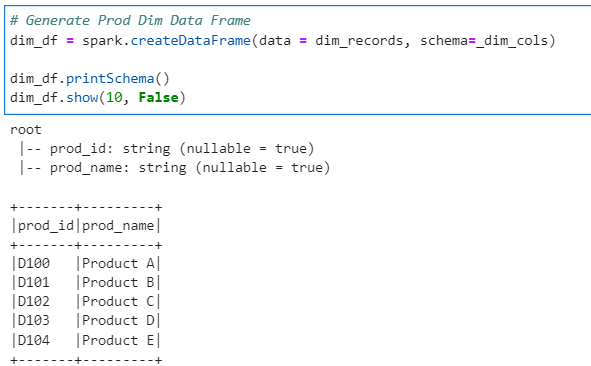
Now, lets set up Spark configuration to disable AQL and set shuffle partitions to 5 for our demonstration
# Set Spark parameters - We have to turn off AQL to demonstrate Salting
spark.conf.set("spark.sql.adaptive.enabled", False)
spark.conf.set("spark.sql.shuffle.partitions", 5)
# Check the parameters
print(spark.conf.get("spark.sql.adaptive.enabled"))
print(spark.conf.get("spark.sql.shuffle.partitions"))
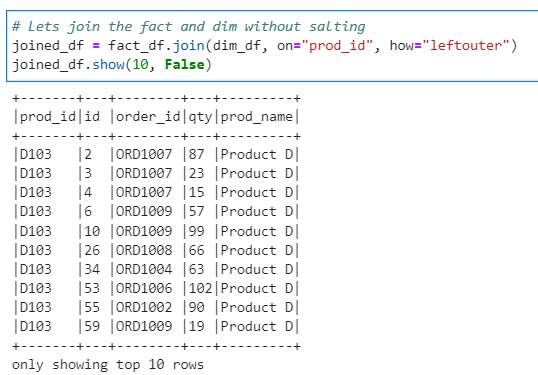
Lets join the Fact and Dim without salting first to check the distribution of data.
# Lets join the fact and dim without salting
joined_df = fact_df.join(dim_df, on="prod_id", how="leftouter")
joined_df.show(10, False)
# Check the partition details to understand distribution
from pyspark.sql.functions import spark_partition_id, count
partition_df = joined_df.withColumn("partition_num", spark_partition_id()).groupBy("partition_num").agg(count("id"))
partition_df.show()
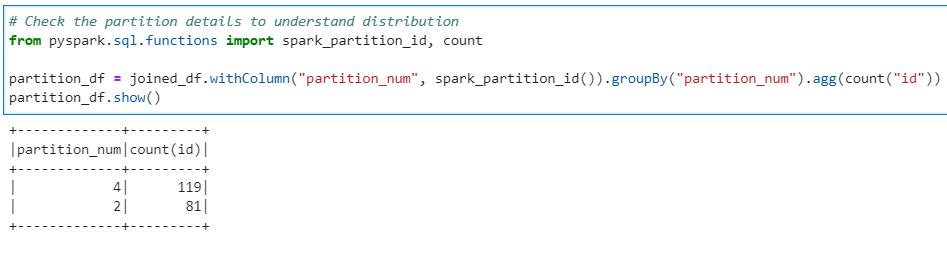
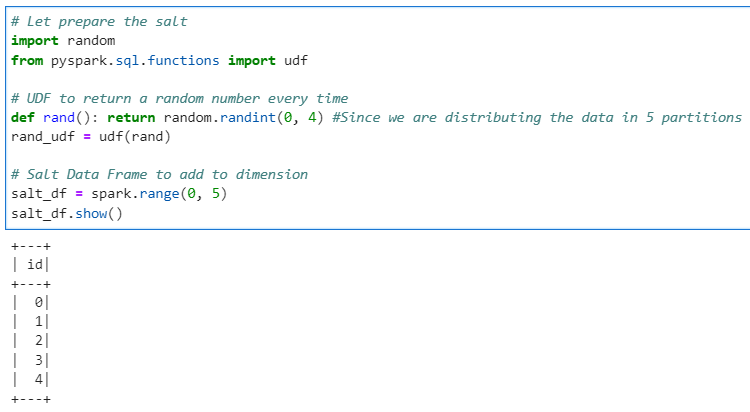
Its very evident that the data is distributed across only 2 partitions out of 5. Now lets generate the salts and salted Fact and Dimension data frames.
# Let prepare the salt
import random
from pyspark.sql.functions import udf
# UDF to return a random number every time
def rand(): return random.randint(0, 4) #Since we are distributing the data in 5 partitions
rand_udf = udf(rand)
# Salt Data Frame to add to dimension
salt_df = spark.range(0, 5)
salt_df.show()
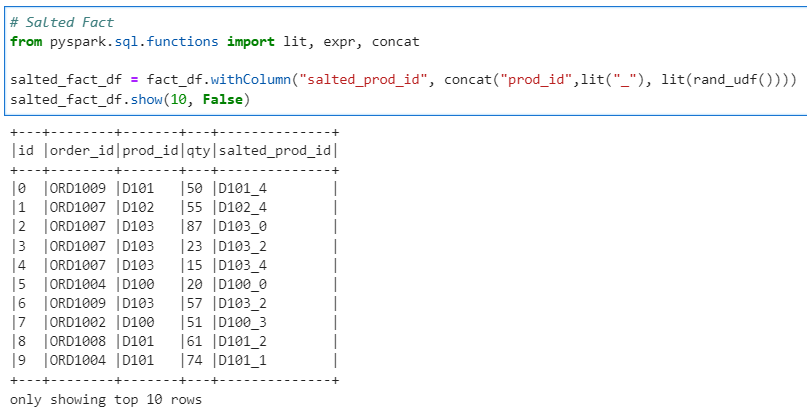
Prepare our salted Fact data frame.
# Salted Fact
from pyspark.sql.functions import lit, expr, concat
salted_fact_df = fact_df.withColumn("salted_prod_id", concat("prod_id",lit("_"), lit(rand_udf())))
salted_fact_df.show(10, False)
And the salted Dim data frame, we need to cross join with salt data frame as to generate all possible salted dimension keys
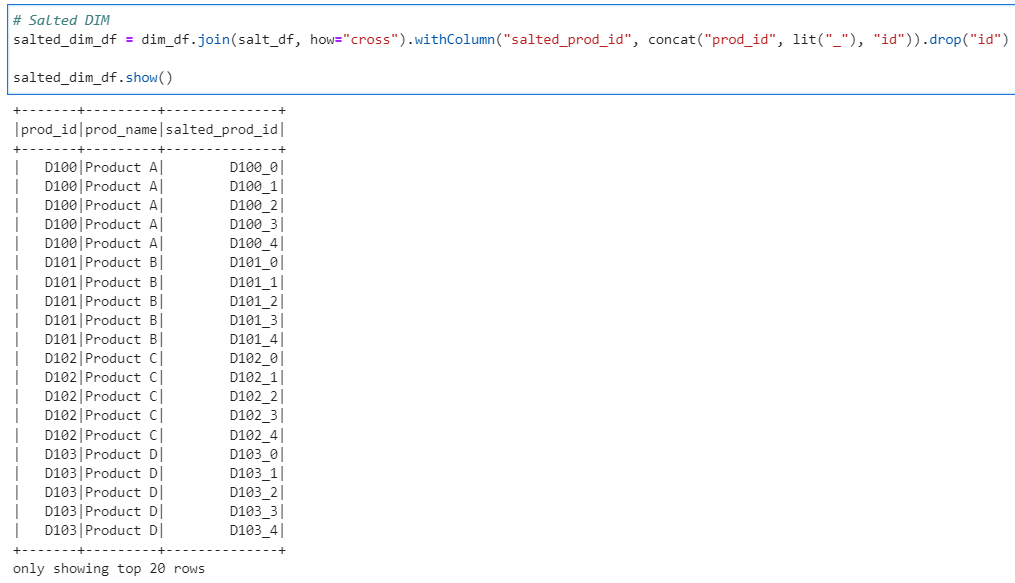
# Salted DIM
salted_dim_df = dim_df.join(salt_df, how="cross").withColumn("salted_prod_id", concat("prod_id", lit("_"), "id")).drop("id")
salted_dim_df.show()
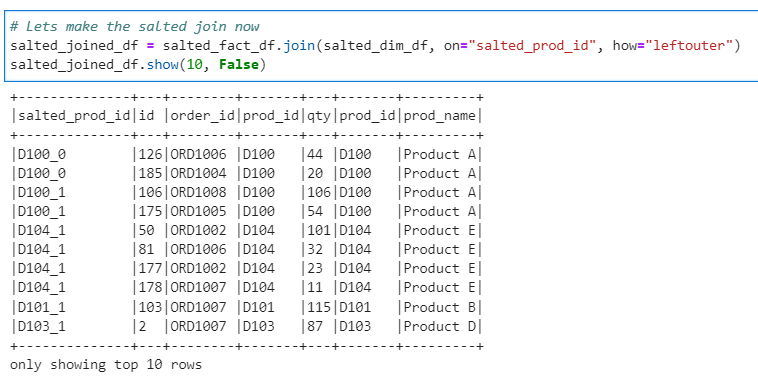
Now, lets join the salted Fact and Dim and check the distribution of data across partitions
# Lets make the salted join now
salted_joined_df = salted_fact_df.join(salted_dim_df, on="salted_prod_id", how="leftouter")
salted_joined_df.show(10, False)
# Check the partition details to understand distribution
from pyspark.sql.functions import spark_partition_id, count
partition_df = salted_joined_df.withColumn("partition_num", spark_partition_id()).groupBy("partition_num") \
.agg(count(lit(1)).alias("count")).orderBy("partition_num")
partition_df.show()
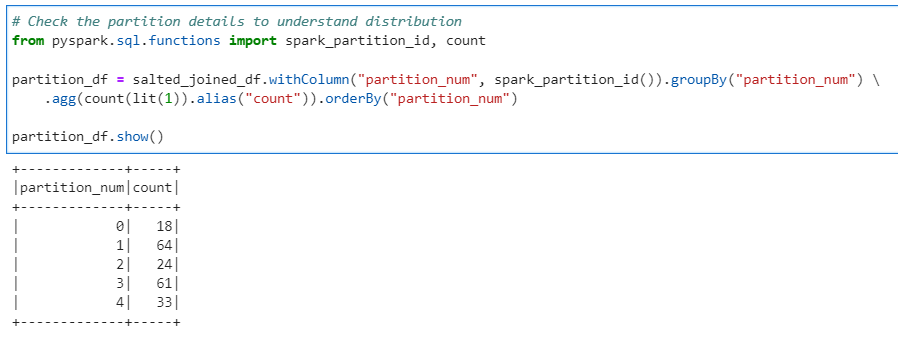
Conclusion: Its evident from the above results that the technique was very effective in distributing data across partitions to avoid data skewness. But, with AQL in place from Spark 3, we don’t need to use salting anymore, as the Adaptive Query Engine take care of the data distribution across partitions effectively.
However, this serves a good learning for us :)
Checkout the iPython Notebook at Github — https://github.com/subhamkharwal/ease-with-apache-spark/blob/master/9_salting_technique.ipynb
Checkout PySpark Series on Medium for more such articles — https://subhamkharwal.medium.com/learnbigdata101-spark-series-940160ff4d30
Top Five
Following are the top five articles as per views. Don't forget check them out:
Buy me a Coffee
If you like my content and wish to buy me a COFFEE. Click the link below or Scan the QR.
Buy Subham a Coffee
*All Payments are secured through Stripe.

About the Author
Subham is working as Senior Data Engineer at a Data Analytics and Artificial Intelligence multinational organization.
Checkout portfolio: Subham Khandelwal